AI-assisted skin cancer detection with imaging data
Background
This project involves building a system to detect skin cancer from lesion images, by training the AI on the
HAM10000 public dataset.
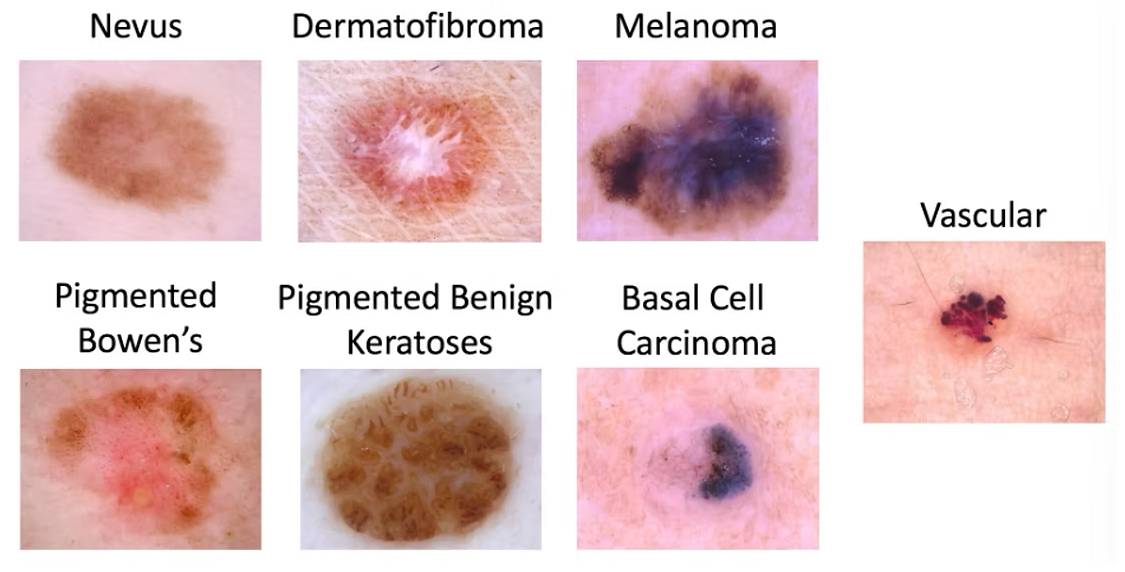
There are 7 types of lesions associated with skin cancer, as seen in the images below.
The dataset contained about 10,000 skin lesion images that fall into 1 of these 7 classes.
The core usecase AI here is to discern if a lesion is potentially cancerous. This can make
detection cheap and accessible, while using AI we can improve accuracy and consistency of the diagnosis.
The goal was to use different AI/ML algorithms to classify lesion images as cancerous or not, evaluate
their effectiveness, and also test if the method has any undesirable properties like data bias.
Implementation
- The implementation used Python notebooks in Google Colab, using the Scikit-learn data science library.
- As with typical AI/ML applications, the data was split 60-40 into training and testing data. The trained model is then used to predict the classification for the unseen testing data to measure accuracy.
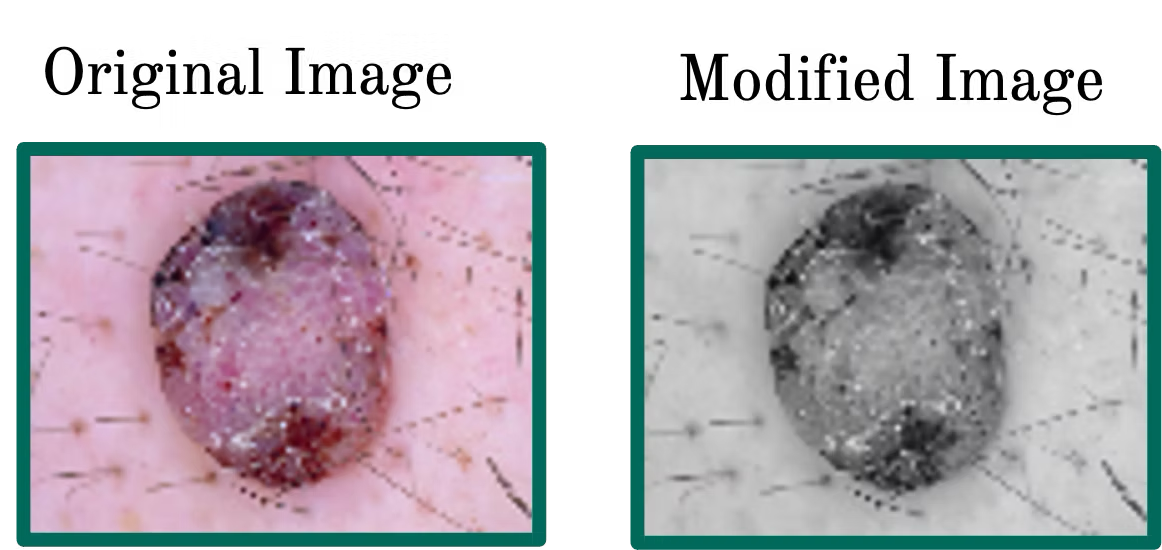
- Feature Engineering prepares and transforms data for model training. This involved turning colored images to grayscale to eliminate effect of skin tone variations, data augmentation and generalization by zooming to eliminate effect of camera position, and blurring the images with a small 4X4 pixel mask, to use softer edges. The images are finally "flattened" from 2D matrices to 1D arrays.
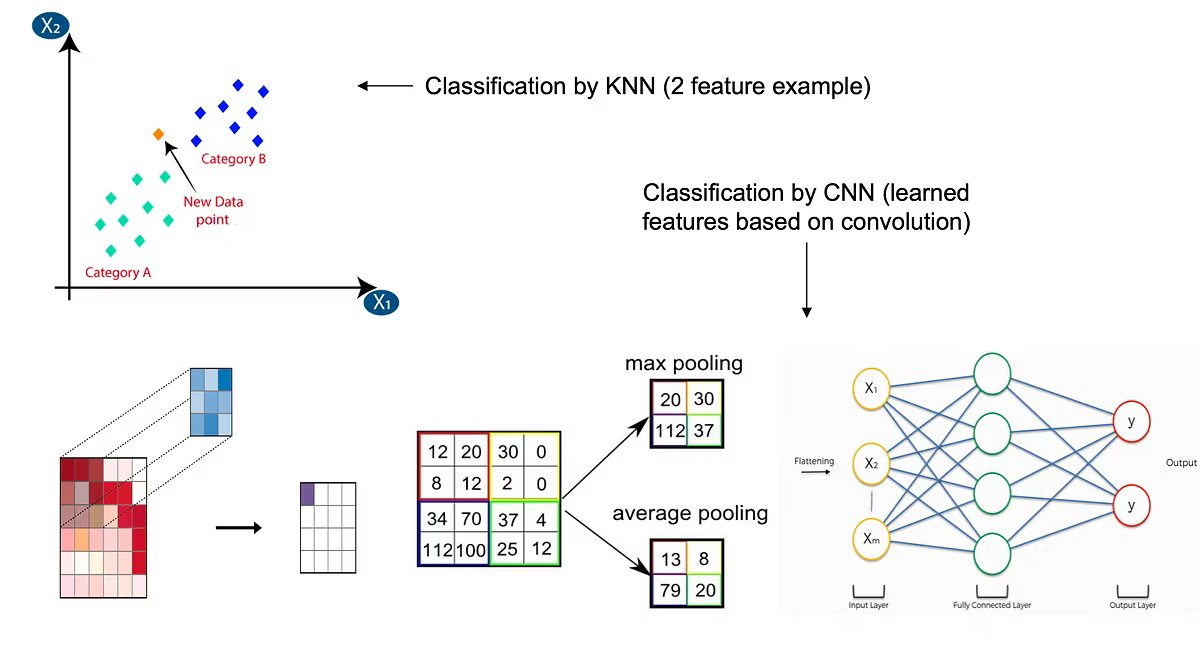
- The 2 algorithms used were K-Nearest-Neighbors (KNN) and Convolutional Neural Networks (CNN). KNN is a classification algorithm that uses "distance" between data points in the feature space to assign a class label based on proximity. CNN learns key features of an image while preserving spatial relationships, and classifies the image to a class based on these features.
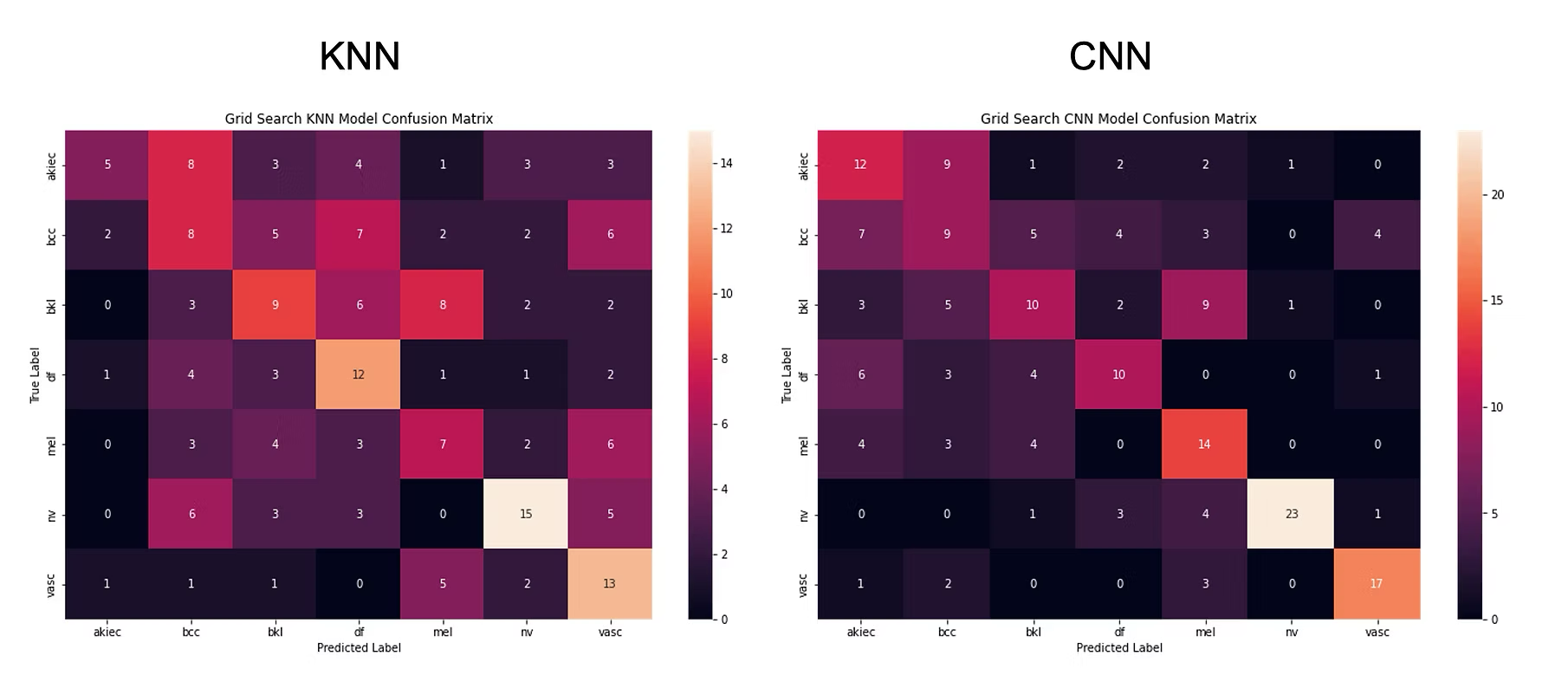
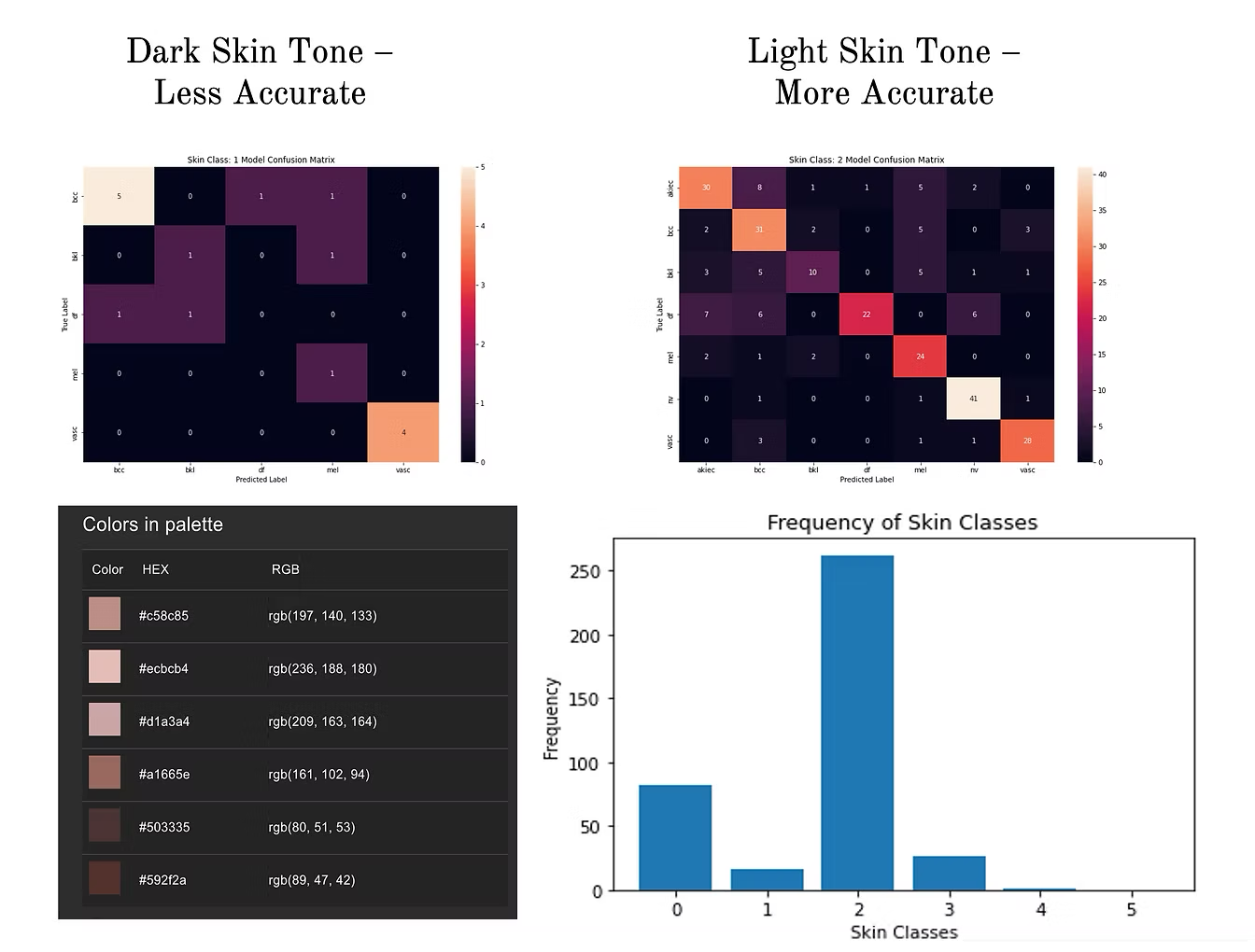
- Effectiveness of these algorithms is assessed using a "Confusion Matrix", capturing the distribution of true vs. predicted labels of the test set. A perfect result will have the matrix "light up" along the diagonal, because it means predicted labels match true labels. Similarly a bad result will have a "scattered" matrix.
- The KNN performs better than CNN, with KNN having 75% accuracy overall and CNN 49%
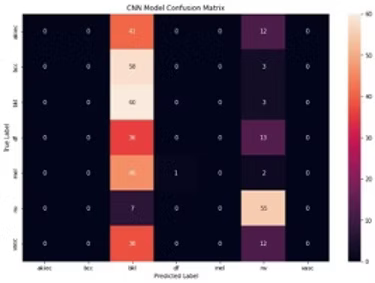
- Hyperparameter selection is critical, which was done through a "Grid Search" that essentially tests the model performance with different parameter settings, and returns the optimal ones. The above results were achieved after applying Grid search to both algorithms. If we don't apply this important step, the performance drops significantly (as shown, a drop to to 29.7% for CNN).
- The final step is to check for bias. The distribution of different skin tones is not uniform in our data set. This is quite common in medical datasets and reflects inequalities in healthcare access by different communities (eg. Caucasian vs. African-American). The algorithms perform better for classes with more samples (e.g. lighter skin tone - class 2), versus classes with lesser data (darker skin tone - class 1). Mitigation of these biases (e.g. by balancing class distributions) is critical for practical application.